Table of Contents
| Table of Contents |
|---|
Profilers
I use two profilers:
cProfile - https://docs.python.org/3/library/profile.html
line-profiler - https://pypi.org/project/line-profiler/
Requirements
python 3.10: module load itm-python/3.10
line-profiler : pip install line-profiler
gprof2dot : pip install gprof2dot
How to run?
- Prepare a standard launch
- Copy
pythondirectory from/gss_efgw_work/work/g2fjc/jintrac/v220922/pythonto some location onpfsfor example:/pfs/work/g2pbloch/python(I use paths with my username, change them to ones with your username) - Change
JINTRAC_PYTHON_DIRin rjettov (430 line) to python directory on pfs from step 2 - Change
run_python_driver (line 49)in/pfs/work/g2pbloch/python(I use paths with my username, change them to ones with your username): - cProfile :
mpirun --allow-run-as-root -np $NPROC python -u -m cProfile -o jintrac.prof /pfs/work/g2pbloch/python/jintrac_imas_driver.py mpi - line-profiler :
mpirun --allow-run-as-root -np $NPROC python -u -m kernprof -l /pfs/work/g2pbloch/python/jintrac_imas_driver.py mpi When we use line-profiler we must add wrapper to profiling function. In this case we should add
@profileupperjintrac_imas_driverfunction injintrac_imas_driver.py:Code Block language py title function wrapper @profile def jintrac_imas_driver(params, components, mpi='no'): """JINTRAC-IMAS generic workflow driver."""- Run
./rjettov -S -I -p -x64 test v220922 g2fjc
cProfile analysis
To read data from test.prof we need python script. We can use e.g.
| Code Block | ||||
|---|---|---|---|---|
| ||||
import pstats
stats = pstats.Stats('test.prof')
stats.strip_dirs().sort_stats(pstats.SortKey.TIME).print_stats() |
This script print data sorted by tottime column ( In tottime column is total time spent in the given function (and excluding time made in calls to sub-functions)). These are the first few lines of the result:
| Code Block |
|---|
Wed Nov 30 09:40:33 2022 test.prof
19379745 function calls (19376063 primitive calls) in 164.514 seconds
Ordered by: internal time
ncalls tottime percall cumtime percall filename:lineno(function)
3 81.304 27.101 81.304 27.101 {imas_3_37_0_ual_4_11_0._ual_lowlevel.ual_open_pulse}
2891220 42.947 0.000 52.709 0.000 {imas_3_37_0_ual_4_11_0._ual_lowlevel.ual_read_data_array}
2497358 16.127 0.000 16.127 0.000 {imas_3_37_0_ual_4_11_0._ual_lowlevel.ual_read_data_scalar}
2899948 2.784 0.000 2.784 0.000 {built-in method numpy.zeros}
815194 2.391 0.000 2.391 0.000 {method 'reduce' of 'numpy.ufunc' objects}
427578 1.225 0.000 2.589 0.000 fromnumeric.py:38(_wrapit)
815196 1.015 0.000 7.988 0.000 {built-in method numpy.core._multiarray_umath.implement_array_function}
387616 0.876 0.000 2.809 0.000 fromnumeric.py:69(_wrapreduction)
427578 0.848 0.000 0.848 0.000 {method 'reshape' of 'numpy.ndarray' objects}
427578 0.685 0.000 3.358 0.000 fromnumeric.py:51(_wrapfunc)
387785 0.684 0.000 0.684 0.000 {method 'items' of 'dict' objects} |
Complete file is below:
| View file | ||||
|---|---|---|---|---|
|
We can also make an image from data:
- Install gprof2dot :
pip install gprof2dot - run :
python -m gprof2dot -f pstats test.prof | dot -Tsvg -o output.svg
Example image looks like this ( output.svg is simplified image, output1.svg is complete image with full data):
View file name output.svg height 250 View file name output1.svg height 250
Line-profiler analysis
To read data from jintrac_imas_driver.py.lprof run:
-
python -m line_profiler jintrac_imas_driver.py.lprof.
These are the first few lines of the results :
| Code Block | ||
|---|---|---|
| ||
Timer unit: 1e-06 s Total time: 232.892 s File: /pfs/work/g2pbloch/python/jintrac_imas_driver.py Function: jintrac_imas_driver at line 721 Line # Hits Time Per Hit % Time Line Contents ============================================================== 721 @profile 722 def jintrac_imas_driver(params, components, mpi='no'): 723 724 """JINTRAC-IMAS generic workflow driver.""" 725 726 # Workflow configuration parameters 727 728 1 5.9 5.9 0.0 user_in = wf.string(params, "Input IDS user") 729 1 2.3 2.3 0.0 machine_in = wf.string(params, "Input IDS machine") 730 1 4.7 4.7 0.0 shot_in = wf.integer(params, "Input IDS shot") 731 1 2.1 2.1 0.0 run_in = wf.integer(params, "Input IDS run") 732 1 2.9 2.9 0.0 user_out = wf.string(params, "Output IDS user") 733 1 2.6 2.6 0.0 machine_out = wf.string(params, "Output IDS machine") 734 1 2.6 2.6 0.0 shot_out = wf.integer(params, "Output IDS shot") 735 1 2.6 2.6 0.0 run_out = wf.integer(params, "Output IDS run") 736 1 13.3 13.3 0.0 user_tmp = os.environ['HOME']+'/public/tempdb' |
Complete file is below:
| View file | ||||
|---|---|---|---|---|
|
Key lines from the results:
| Code Block | ||
|---|---|---|
| ||
Line # Hits Time Per Hit % Time Line Contents
895 1 62545203.7 62545203.7 26.9 alenv = ALEnv(user_temp=user_tmp)
917 22 49496025.7 2249819.4 21.3 ids_bundle_input[ids_struct] = eval('DBentry.idsin.get("'+ids_struct+'")')
922 1 83745046.9 83745046.9 36.0 tmpdict = bundle_copy(ids_bundle_input)
956 2 4665617.1 2332808.6 2.0 ids_bundle_input[elem] = DBentry.idsin.get_slice(elem, tstart, 3)
966 1 6353190.5 6353190.5 2.7 ids_bundle_work = bundle_copy(ids_bundle_input)
967 1 7884627.8 7884627.8 3.4 ids_bundle_updated = bundle_copy(ids_bundle_input)
1022 3 14539266.0 4846422.0 6.2 ids_bundle_prev[item] = bundle_copy(ids_bundle_work,imas_control.get_ids_sublist_updates(item)) |
Candidates for bottlenecks
From cProfile result can see that 85% execution time jintrac_imas_driver.py spend on imas functions. Function ual_open_pulse was called only 3 times. It execution time is 27s. Functions ual_read_data_array and ual_read_data_scalar was called more than 2 milions times.
From line-profiler we can see, that jintrac_imas_driver.py spends a lot of time in lins with functioin bundle_copy. This function call imas functions. A lot of time is also taken up by functions calls with DBentry.
Possible ways to speed up jintrac_imas_driver.py:
Reducing number of calls: ual_open_pulse, ual_read_data_array, ual_read_data_scalar
Summary of July Profiling (HFD5)
Instruction
| Code Block | ||
|---|---|---|
| ||
module use /pfs/work/g2fjc/jintrac/v120523/modules module load jintrac export JINTRAC_IMAS_BACKEND=HDF5 module use /pfs/g2fjc/cmg/jams/v230123/modules module load jams (Create test case data) idscp -u g2pmanas --database west -si 54762 -so 54762 -ri 123 -ro 123 -bo HDF5 (path) /pfs/work/g2fjc/jetto/runs/runmemleak_v120523_np1_z0_1ms |
Line Profiler
Key lines from the results:
| Code Block | ||
|---|---|---|
| ||
Timer unit: 1e-06 s
Total time: 155.404 s
File: /pfs/work/g2pbloch/jintrac/python/jintrac_imas_driver.py
Function: jintrac_imas_driver at line 735
Line # Hits Time Per Hit % Time Line Contents
==============================================================
957 22 44253622.5 2011528.3 28.5 ids_bundle_input[ids_struct] = eval('DBentry.idsin.get("'+ids_struct+'")')
962 1 46906374.5 46906374.5 30.2 tmpdict = bundle_copy(ids_bundle_input)
996 2 5116586.7 2558293.3 3.3 ids_bundle_input[elem] = DBentry.idsin.get_slice(elem, tstart, interp_value)
1011 1 3447098.9 3447098.9 2.2 ids_bundle_work = bundle_copy(ids_bundle_input)
1012 1 4400338.9 4400338.9 2.8 ids_bundle_updated = bundle_copy(ids_bundle_input)
1067 3 6886544.4 2295514.8 4.4 ids_bundle_prev[item] = bundle_copy(ids_bundle_work,imas_control.get_ids_sublist_updates(item))
1099 3 29669430.5 9889810.2 19.1 ids_bundle_updated_tmp = eval(imas_control.get_wrapper(item) + args)
1105 11 4863627.8 442148.0 3.1 ids_bundle_updated[ids_struct].copyValues(ids_bundle_updated_tmp[ids_struct])
1106 11 3643527.1 331229.7 2.3 ids_bundle_prev[item][ids_struct].copyValues(ids_bundle_updated_tmp[ids_struct])
1114 1 4798639.4 4798639.4 3.1 ids_bundle_work = bundle_copy(ids_bundle_updated)
Total time: 12.4112 s
File: /pfs/work/g2pbloch/jintrac/python/jintrac_imas_driver.py
Function: jetto_wrapper at line 304
Line # Hits Time Per Hit % Time Line Contents
==============================================================
323 1 0.7 0.7 0.0 out_prof, out_src, out_trans, out_equil, out_summ, out_num, out_work \
324 1 8768238.1 8768238.1 70.6 = jetto(in_prof, in_src, in_trans, in_equil, in_nbi,
325 1 0.3 0.3 0.0 in_pel, in_pulse, in_contr, in_summ, in_num, in_work)
367 1 3589306.4 3589306.4 28.9 out_ids_bundle = bundle_copy(in_ids_bundle_work) |
Complete file is below:
| View file | ||||
|---|---|---|---|---|
|
cProfiler
| Code Block | ||
|---|---|---|
| ||
Wed Jul 5 09:31:02 2023 jintrac.prof
207387232 function calls (178651380 primitive calls) in 151.148 seconds
Ordered by: internal time
ncalls tottime percall cumtime percall filename:lineno(function)
26076870/927 40.102 0.000 82.859 0.089 copy.py:128(deepcopy)
3356451 24.272 0.000 36.639 0.000 {imas_3_38_1_ual_4_11_4._ual_lowlevel.ual_read_data_array}
778014/927 9.056 0.000 82.841 0.089 copy.py:227(_deepcopy_dict)
53709341 7.111 0.000 7.111 0.000 {method 'get' of 'dict' objects}
1 5.916 5.916 7.367 7.367 wrapper.py:48(jetto_actor)
2959395 4.441 0.000 4.441 0.000 {imas_3_38_1_ual_4_11_4._ual_lowlevel.ual_read_data_scalar}
4441187 4.234 0.000 4.234 0.000 {method '__deepcopy__' of 'numpy.ndarray' objects}
1556028/927 4.081 0.000 82.851 0.089 copy.py:259(_reconstruct)
7099231 3.948 0.000 5.721 0.000 copy.py:243(_keep_alive)
35834998 3.774 0.000 3.774 0.000 {built-in method builtins.id}
3398693 3.110 0.000 3.110 0.000 {built-in method numpy.zeros}
1005851 3.045 0.000 3.045 0.000 {method 'reduce' of 'numpy.ufunc' objects}
18974180 2.106 0.000 2.106 0.000 copy.py:182(_deepcopy_atomic)
1556028 1.918 0.000 1.921 0.000 {method '__reduce_ex__' of 'object' objects}
8578377 1.660 0.000 1.660 0.000 {built-in method builtins.getattr}
134159 1.607 0.000 1.607 0.000 {imas_3_38_1_ual_4_11_4._ual_lowlevel.ual_begin_arraystruct_action}
509900 1.504 0.000 3.231 0.000 fromnumeric.py:38(_wrapit)
1005853 1.383 0.000 10.248 0.000 {built-in method numpy.core._multiarray_umath.implement_array_function}
1 1.212 1.212 151.202 151.202 jintrac_imas_driver.py:1(<module>)
1274146 1.140 0.000 1.140 0.000 {method 'items' of 'dict' objects}
495951 1.116 0.000 3.696 0.000 fromnumeric.py:69(_wrapreduction)
509900 1.051 0.000 1.051 0.000 {method 'reshape' of 'numpy.ndarray' objects} |
Complete file is below:
View file name cprofile_result.txt height 250
Possible bottlenecks
- Process of copying data (
bundle_copy,copyValues). eval(imas_control.get_wrapper(item) + args)andeval(imas_control.get_wrapper(item) + args)take almost half of function time
Testing HDF5
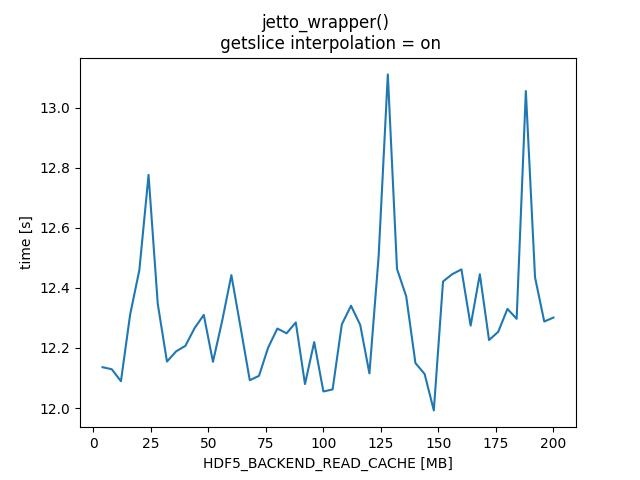
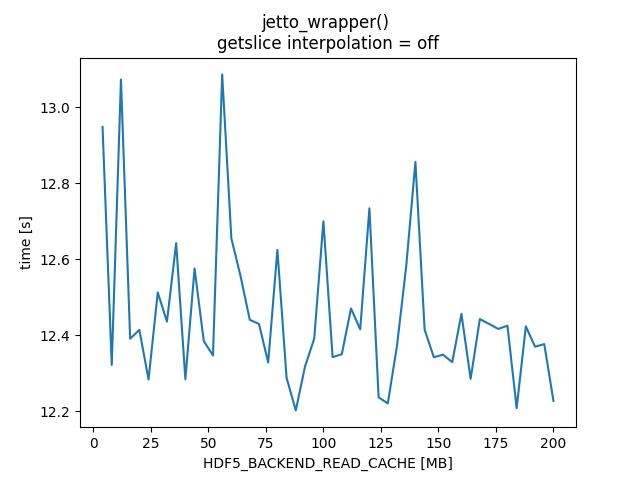
HDF5_BACKEND_READ_CACHE - TABLE
| Markdown |
|---|
|HDF5_BACKEND_READ_CACHE [MB] |jintrac_imas_driver |jetto_wrapper |jintrac_imas_driver |jetto_wrapper| |getslice interpolation| off | off| on |on | |----------|------------|------------|-----------|-------------| | 4 | 241.739 | 12.9481 | 230.008 | 12.136 | | 8 | 234.774 | 12.3213 | 228.837 | 12.1292 | | 12 | 168.333 | 13.0729 | 148.889 | 12.0893 | | 16 | 154.656 | 12.3905 | 152.87 | 12.3117 | | 20 | 152.988 | 12.4138 | 155.945 | 12.4595 | | 24 | 150.913 | 12.283 | 155.469 | 12.7765 | | 28 | 153.991 | 12.5123 | 153.11 | 12.3468 | | 32 | 153.903 | 12.4356 | 149.575 | 12.1548 | | 36 | 159.225 | 12.6422 | 151.492 | 12.1888 | | 40 | 149.549 | 12.2838 | 152.926 | 12.207 | | 44 | 158.385 | 12.5754 | 154.827 | 12.266 | | 48 | 153.306 | 12.3843 | 152.786 | 12.3101 | | 52 | 150.771 | 12.3464 | 151.305 | 12.1539 | | 56 | 165.846 | 13.0865 | 153.383 | 12.2905 | | 60 | 153.729 | 12.6542 | 155.563 | 12.4427 | | 64 | 161.231 | 12.5552 | 151.229 | 12.2685 | | 68 | 154.142 | 12.4402 | 148.697 | 12.0926 | | 72 | 153.52 | 12.4294 | 149.757 | 12.1072 | | 76 | 149.982 | 12.3277 | 151.765 | 12.2017 | | 80 | 155.473 | 12.6245 | 151.903 | 12.2647 | | 84 | 150.118 | 12.2872 | 152.894 | 12.2489 | | 88 | 149.918 | 12.2017 | 154.456 | 12.285 | | 92 | 151.403 | 12.317 | 149.207 | 12.0798 | | 96 | 150.825 | 12.3905 | 152.792 | 12.2197 | | 100 | 149.834 | 12.6997 | 147.859 | 12.0551 | | 104 | 150.326 | 12.3421 | 148.031 | 12.062 | | 108 | 151.51 | 12.3496 | 152.049 | 12.2785 | | 112 | 155.506 | 12.4702 | 154.526 | 12.3407 | | 116 | 153.662 | 12.4152 | 153.498 | 12.2769 | | 120 | 158.311 | 12.7339 | 148.607 | 12.1153 | | 124 | 154.864 | 12.2361 | 156.592 | 12.5044 | | 128 | 153.5 | 12.2201 | 154.231 | 13.1112 | | 132 | 152.191 | 12.3723 | 156.739 | 12.4632 | | 136 | 160.911 | 12.5815 | 152.368 | 12.3718 | | 140 | 159.165 | 12.8561 | 149.531 | 12.1498 | | 144 | 153.646 | 12.4145 | 151.239 | 12.1133 | | 148 | 149.434 | 12.3417 | 147.887 | 11.9921 | | 152 | 151.505 | 12.3484 | 151.848 | 12.4215 | | 156 | 151.068 | 12.3288 | 156.421 | 12.4458 | | 160 | 152.951 | 12.4558 | 155.213 | 12.4617 | | 164 | 149.566 | 12.2848 | 151.589 | 12.2744 | | 168 | 151.872 | 12.4422 | 151.654 | 12.4456 | | 172 | 152.766 | 12.4294 | 153.34 | 12.2264 | | 176 | 151.982 | 12.4162 | 149.911 | 12.254 | | 180 | 152.768 | 12.4248 | 156.015 | 12.3302 | | 184 | 148.166 | 12.2076 | 149.323 | 12.2971 | | 188 | 153.493 | 12.4229 | 153.415 | 13.0557 | | 192 | 153.968 | 12.3694 | 157.029 | 12.4348 | | 196 | 152.465 | 12.3764 | 154.288 | 12.2881 | | 200 | 149.238 | 12.2266 | 154.713 | 12.3013 | |
HDF5_BACKEND_READ_CACHE - PLOTS
TESTING MDSPLUS
LINE PROFILER
| Code Block | ||
|---|---|---|
| ||
Timer unit: 1e-06 s
Total time: 155.743 s
File: /pfs/work/g2pbloch/jintrac/python/jintrac_imas_driver.py
Function: jintrac_imas_driver at line 735
Line # Hits Time Per Hit % Time Line Contents
==============================================================
896 1 1638364.1 1638364.1 1.1 status, idx_out = DBentry.idsout.create() # idx_out points to output IDS file index
957 22 50742304.5 2306468.4 32.6 ids_bundle_input[ids_struct] = eval('DBentry.idsin.get("'+ids_struct+'")')
962 1 45878227.2 45878227.2 29.5 tmpdict = bundle_copy(ids_bundle_input)
996 2 3193050.2 1596525.1 2.1 ids_bundle_input[elem] = DBentry.idsin.get_slice(elem, tstart, interp_value)
1011 1 3399663.8 3399663.8 2.2 ids_bundle_work = bundle_copy(ids_bundle_input)
1012 1 4340984.3 4340984.3 2.8 ids_bundle_updated = bundle_copy(ids_bundle_input)
1067 3 6779104.9 2259701.6 4.4 ids_bundle_prev[item] = bundle_copy(ids_bundle_work,imas_control.get_ids_sublist_updates(item))
1199 3 25454536.5 8484845.5 16.3 ids_bundle_updated_tmp = eval(imas_control.get_wrapper(item) + args)
1105 11 4817487.6 437953.4 3.1 ids_bundle_updated[ids_struct].copyValues(ids_bundle_updated_tmp[ids_struct])
1106 11 3587909.3 326173.6 2.3 ids_bundle_prev[item][ids_struct].copyValues(ids_bundle_updated_tmp[ids_struct])
1114 1 4769085.3 4769085.3 3.1 ids_bundle_work = bundle_copy(ids_bundle_updated)
Total time: 12.4592 s
File: /pfs/work/g2pbloch/jintrac/python/jintrac_imas_driver.py
Function: jetto_wrapper at line 304
Line # Hits Time Per Hit % Time Line Contents
==============================================================
323 1 1.3 1.3 0.0 out_prof, out_src, out_trans, out_equil, out_summ, out_num, out_work \
324 1 8958798.8 8958798.8 71.9 = jetto(in_prof, in_src, in_trans, in_equil, in_nbi,
325 1 0.2 0.2 0.0 in_pel, in_pulse, in_contr, in_summ, in_num, in_work)
367 1 3459717.4 3459717.4 27.8 out_ids_bundle = bundle_copy(in_ids_bundle_work)
|
Complete file is below:
View file name line_profiler_result.txt height 250
CPROFILER
| Code Block | ||
|---|---|---|
| ||
Thu Jul 20 14:45:51 2023 jintrac.prof
207169007 function calls (178433155 primitive calls) in 153.082 seconds
Ordered by: internal time
ncalls tottime percall cumtime percall filename:lineno(function)
26076870/927 40.886 0.000 83.306 0.090 copy.py:128(deepcopy)
3356451 22.884 0.000 35.138 0.000 {imas_3_38_1_ual_4_11_3._ual_lowlevel.ual_read_data_array}
778014/927 8.909 0.000 83.288 0.090 copy.py:227(_deepcopy_dict)
2959395 8.181 0.000 8.181 0.000 {imas_3_38_1_ual_4_11_3._ual_lowlevel.ual_read_data_scalar}
53709341 6.903 0.000 6.903 0.000 {method 'get' of 'dict' objects}
1 5.946 5.946 7.387 7.387 wrapper.py:48(jetto_actor)
4441187 4.289 0.000 4.289 0.000 {method '__deepcopy__' of 'numpy.ndarray' objects}
1556028/927 4.099 0.000 83.298 0.090 copy.py:259(_reconstruct)
7099231 3.986 0.000 5.677 0.000 copy.py:243(_keep_alive)
35834998 3.833 0.000 3.833 0.000 {built-in method builtins.id}
3398693 2.945 0.000 2.945 0.000 {built-in method numpy.zeros}
987875 2.815 0.000 2.815 0.000 {method 'reduce' of 'numpy.ufunc' objects}
18974180 2.163 0.000 2.163 0.000 copy.py:182(_deepcopy_atomic)
1556028 1.704 0.000 1.707 0.000 {method '__reduce_ex__' of 'object' objects}
8542427 1.672 0.000 1.672 0.000 {built-in method builtins.getattr}
495951 1.465 0.000 4.334 0.000 fromnumeric.py:69(_wrapreduction)
1274146 1.441 0.000 1.441 0.000 {method 'items' of 'dict' objects}
491924 1.422 0.000 3.040 0.000 fromnumeric.py:38(_wrapit)
1 1.277 1.277 153.141 153.141 jintrac_imas_driver.py:1(<module>)
987877 1.196 0.000 10.454 0.000 {built-in method numpy.core._multiarray_umath.implement_array_function}
491924 1.001 0.000 1.001 0.000 {method 'reshape' of 'numpy.ndarray' objects} |
Complete file is below:
View file name cprofile_result.txt height 250
Partial_get vs get
IDS :: Core_profiles
The test is checking the time differences between fetching whole IDS using get and fetching IDS in parts using partial_get .
| Code Block | ||||
|---|---|---|---|---|
| ||||
import sys
import numpy
import os
import time
import imas
user = os.environ['USER']
imasVersion = os.environ['IMAS_VERSION'][:1]
database = 'test'
backendtype = 'HDF5'
machine_in = "west"
ids_in = imas.DBEntry(13, "west", 54762, 1, user_name="/pfs/work/g2pbloch/jetto/runs/test/imasdb")
ids_in.open()
start_partial = time.time()
rho_tor_norm = ids_in.partial_get("core_profiles" ,"profiles_1d(:)/grid/rho_tor_norm(:)")
core_profiles_profiles_1d_zeff = ids_in.partial_get("core_profiles", "profiles_1d(:)/zeff(:)")
rho_tor_norm = ids_in.partial_get("core_profiles" ,"profiles_1d(:)/grid/rho_tor_norm(:)")
q = ids_in.partial_get("core_profiles" ,"profiles_1d(:)/q(:)")
density_thermal = ids_in.partial_get("core_profiles" ,"profiles_1d(:)/electrons.density_thermal(:)")
temperature = ids_in.partial_get("core_profiles" ,"profiles_1d(:)/electrons/temperature(:)")
ion = ids_in.partial_get("core_profiles" ,"profiles_1d(:)/ion")
t_i_average = ids_in.partial_get("core_profiles" ,"profiles_1d(:)/t_i_average(:)")
zeff = ids_in.partial_get("core_profiles" ,"profiles_1d(:)/zeff(:)")
rotation_frequency_tor_sonic = ids_in.partial_get("core_profiles", "profiles_1d(:)/rotation_frequency_tor_sonic")
end_partial = time.time()
start_get = time.time()
core_profiles = ids_in.get("core_profiles")
end_get = time.time()
print(f"partial_get: {end_partial - start_partial}, get : { end_get - start_get}")
ids_in.close() |
The time gain is small, just below 1 second, for the entire duration of about 24 seconds:
partial_get: 23.91968011856079, get : 24.872377157211304
IDS :: equilibrium
Testing description
I couldn't retrieve the data using, for example: equilibriumtime_sliceitimeglobal_quantitiesip = ids_in.partial_get("equilibrium", "time_slice(:)/global_quantities/ip") because I was getting the following error: ERROR: [time_slice] AoS internal field(s) cannot be returned in non-homogeneous mode for more than one AoS index . Due to this, I had to iterate and retrieve the data for each time separately.
When it comes to get_slice , I retrieve timestamps using partial_get . Then, for each timestamp, I fetch a segment using get_slice and apply imas.imasdef.LINEAR_INTERP interpolation.
| Code Block | ||||
|---|---|---|---|---|
| ||||
import sys
import numpy
import os
import time
import imas
def run():
user = os.environ['USER']
imasVersion = os.environ['IMAS_VERSION'][:1]
database = 'test'
backendtype = 'HDF5'
machine_in = "west"
ids_in = imas.DBEntry(13, "west", 54762, 1, user_name="/pfs/work/g2pbloch/jetto/runs/test/imasdb")
ids_in.open()
start_partial = time.time()
eq_time = ids_in.partial_get("equilibrium", "time(:)")
equilibriumtime_sliceitimeglobal_quantitiesip = [ids_in.partial_get("equilibrium", f"time_slice({i})/global_quantities/ip") for i in range(len(eq_time))]
equilibriumtime_sliceitimeglobal_quantitiespsi_axis = [ids_in.partial_get("equilibrium", f"time_slice({i})/global_quantities/psi_axis") for i in range(len(eq_time))]
equilibriumtime_sliceitimeglobal_quantitiespsi_boundary = [ids_in.partial_get("equilibrium", f"time_slice({i})/global_quantities/psi_boundary") for i in range(len(eq_time))]
equilibriumtime_sliceitimeglobal_quantitiesmagnetic_axisr = [ids_in.partial_get("equilibrium", f"time_slice({i})/global_quantities/magnetic_axis/r") for i in range(len(eq_time))]
equilibriumtime_sliceitimeglobal_quantitiesmagnetic_axisz = [ids_in.partial_get("equilibrium", f"time_slice({i})/global_quantities/magnetic_axis/z") for i in range(len(eq_time))]
equilibriumtime_sliceitimeprofiles_1drho_tor_norm = [ids_in.partial_get("equilibrium", f"time_slice({i})/profiles_1d/rho_tor_norm(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeprofiles_1dpsi = [ids_in.partial_get("equilibrium", f"time_slice({i})/profiles_1d/psi(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeboundaryoutliner = [ids_in.partial_get("equilibrium", f"time_slice({i})/boundary/outline/r(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeboundarylcfsr = [ids_in.partial_get("equilibrium", f"time_slice({i})/boundary/lcfs/r(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeboundaryoutlinez = [ids_in.partial_get("equilibrium", f"time_slice({i})/boundary/outline/z(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeboundarylcfsz = [ids_in.partial_get("equilibrium", f"time_slice({i})/boundary/lcfs/z(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeprofiles_2d1psi = [ids_in.partial_get("equilibrium", f"time_slice({i})/profiles_2d(1)/psi(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeprofiles_2d1griddim1 = [ids_in.partial_get("equilibrium", f"time_slice({i})/profiles_2d(1)/grid/dim1(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeprofiles_2d1griddim2 = [ids_in.partial_get("equilibrium", f"time_slice({i})/profiles_2d(1)/grid/dim2(:)") for i in range(len(eq_time))]
end_partial = time.time()
start_get = time.time()
equilibrium = ids_in.get("equilibrium")
end_get = time.time()
print(f"partial_get: {end_partial - start_partial}, get : {end_get - start_get}")
ids_in.close()
if __name__ == "__main__":
run() |
| Code Block | ||||
|---|---|---|---|---|
| ||||
import sys
import numpy
import os
import time
import imas
def run():
user = os.environ['USER']
imasVersion = os.environ['IMAS_VERSION'][:1]
database = 'test'
backendtype = 'HDF5'
machine_in = "west"
ids_in = imas.DBEntry(13, "west", 54762, 1, user_name="/pfs/work/g2pbloch/jetto/runs/test/imasdb")
ids_in.open()
eq_time = ids_in.partial_get("equilibrium", "time(:)")
get_slice_start = time.time()
data = [ids_in.get_slice("equilibrium", t, imas.imasdef.LINEAR_INTERP) for t in eq_time]
get_slice_end = time.time()
print(get_slice_end-get_slice_start)
if __name__ == "__main__":
run() |
Time result
- partial_get without
profiles_2d: 4.72s - partial_get with
profiles_2d: 9.682 (profiles_2d(1)/psi: 3.6s ,profiles_2d(1)/grid/dim1: 0.76s ,profiles_2d(1)/grid/dim2: 0.7s)
- get : 7.47s
- get_slice : 718 s
September 2024
Partial_get vs get
environment:
| Code Block | ||||
|---|---|---|---|---|
| ||||
module use /pfs/work/g2fjc/jintrac/32.0.1/modules/
module load jintrac
export JINTRAC_IMAS_BACKEND=HDF5
module use /pfs/work/g2fjc/cmg/jams/31.0.0/modules
module load jams |
Timing (1D) and (1D+2D) with partial get vs full get. I am using data from /gss_efgw_work/work/g2fjc/jetto/runs/runmemleak_v120523_np1_z0_1ms_hdf5/imasdb/ .
Python code to reproduce:
| Code Block | ||||
|---|---|---|---|---|
| ||||
import sys
import numpy
import os
import time
import imas
def run():
user = os.environ['USER']
imasVersion = os.environ['IMAS_VERSION'][:1]
database = 'test'
backendtype = 'HDF5'
machine_in = "west"
ids_in = imas.DBEntry(13, "west", 54762, 1, user_name="/pfs/work/g2pbloch/jetto/runs/test/imasdb")
ids_in.open()
start_partial = time.time()
eq_time = ids_in.partial_get("equilibrium", "time(:)")
equilibriumtime_sliceitimeglobal_quantitiesip = [ids_in.partial_get("equilibrium", f"time_slice({i})/global_quantities/ip") for i in range(len(eq_time))]
equilibriumtime_sliceitimeglobal_quantitiespsi_axis = [ids_in.partial_get("equilibrium", f"time_slice({i})/global_quantities/psi_axis") for i in range(len(eq_time))]
equilibriumtime_sliceitimeglobal_quantitiespsi_boundary = [ids_in.partial_get("equilibrium", f"time_slice({i})/global_quantities/psi_boundary") for i in range(len(eq_time))]
equilibriumtime_sliceitimeglobal_quantitiesmagnetic_axisr = [ids_in.partial_get("equilibrium", f"time_slice({i})/global_quantities/magnetic_axis/r") for i in range(len(eq_time))]
equilibriumtime_sliceitimeglobal_quantitiesmagnetic_axisz = [ids_in.partial_get("equilibrium", f"time_slice({i})/global_quantities/magnetic_axis/z") for i in range(len(eq_time))]
equilibriumtime_sliceitimeprofiles_1drho_tor_norm = [ids_in.partial_get("equilibrium", f"time_slice({i})/profiles_1d/rho_tor_norm(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeprofiles_1dpsi = [ids_in.partial_get("equilibrium", f"time_slice({i})/profiles_1d/psi(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeboundaryoutliner = [ids_in.partial_get("equilibrium", f"time_slice({i})/boundary/outline/r(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeboundaryoutlinez = [ids_in.partial_get("equilibrium", f"time_slice({i})/boundary/outline/z(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeprofiles_2d1psi = [ids_in.partial_get("equilibrium", f"time_slice({i})/profiles_2d(1)/psi(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeprofiles_2d1griddim1 = [ids_in.partial_get("equilibrium", f"time_slice({i})/profiles_2d(1)/grid/dim1(:)") for i in range(len(eq_time))]
equilibriumtime_sliceitimeprofiles_2d1griddim2 = [ids_in.partial_get("equilibrium", f"time_slice({i})/profiles_2d(1)/grid/dim2(:)") for i in range(len(eq_time))]
end_partial = time.time()
start_get = time.time()
equilibrium = ids_in.get("equilibrium")
end_get = time.time()
print(f"partial_get: {end_partial - start_partial}, get : {end_get - start_get}")
ids_in.close()
if __name__ == "__main__":
run()
|
The scientific work is published for the realization of the international project co-financed by Polish Ministry of Science and Higher Education in 2019 from financial resources of the program entitled "PMW"; Agreement No. 5040/H2020/Euratom/2019/2
This work has been carried out within the framework of the EUROfusion Consortium and has received funding from the Euratom research and training programme 2014–2020 under grant agreement No 633053. The views and opinions expressed herein do not necessarily reflect those of the European Commission or ITER